| > DNA Methylation analysis PCR Primer Database |
- How to query the database?
- How to interpret the search results?
- How to register?
- How to submit data?
- How to submit large amounts of data?
- Which fields need special attention for data submission?
- How to change your registration properties?
- How to construct a link to a specific record?
- How to quote/cite the use of primers from the methPrimerDB database?
- What's the software and hardware behind this database?
| How to query the database? |
It's really simple. Just click on 'Search' and confine your search in the form by choosing one of the available methods or organisms. More specifically, you can search for a gene symbol/name, Entrez Gene ID, PubMed ID from an article in which the primer sequences are described, primer sequence or submitter's name. By default, when you query for a gene name or gene symbol, all records that match the substring are retrieved. We discourage the use of alias symbols (e.g. G3PD instead of GAPD for human glyceraldehyde-3-phosphate dehydrogenase), but it's still possible to use them for a query. It's possible to restrict your search by looking for the exact phrase. Last but not least, one can search for a record by a specific methPrimerDB ID.
If you want to order your search results, choose one of the possibilities in the 'Order By' field.
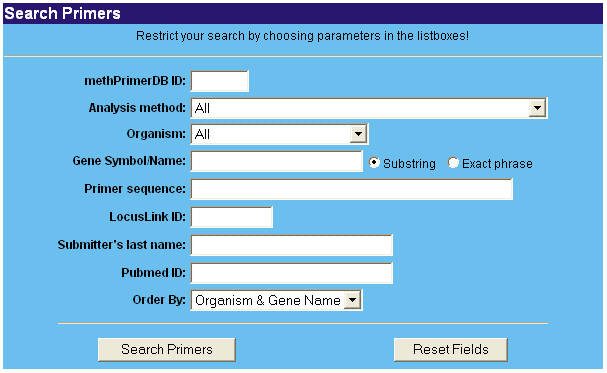
Alternatively, you can perform a quick search on the homepage. Here, you can only search by gene name or gene symbol (substring or exact phrase) or by methPrimerDB ID. It's possible to restrict the search to one organism.

Press 'Enter' or click 'Search' to send your request and obtain the search results.
| How to interpret the search results? |
Result page
The data on the result page depends on the number and the type of hits found in the database.
- First, if you used a keyword which has no match with a record (e.g. a non-existing gene symbol like ABCDEF), you see a warning message of the following type:
No primer/probe records found in the methPrimerDB for the string: ABCDEF
- Second, you are looking for an existing gene for which no primer/probe records are available in the database (yet). Your search will therefore give an overview of all the genes (available in Entrez Gene) of which the gene name, gene symbol or alias symbols partly match with the search string.
Click on the gene symbol to go directly to the Entrez Gene record for this gene. You're kindly invited to submit your primer/probe sequences for one of the listed genes.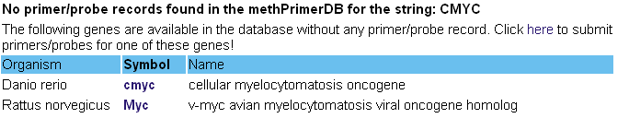
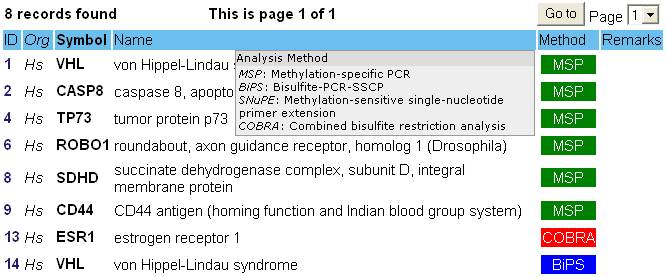
- Third, your search was succesful and you get a tabulated overview of the search results.
The first column lists the methPrimerDB ID. By clicking on this number you can go to the assay report of the primer set. The second column contains the abbreviation of the organism for which record was submitted. In the third and fourth column, the gene symbol and the gene name are displayed.
In the next column ('Method'), the method of the primer record is shown with an abbreviation and a colored box. To know the meaning of the abbreviation, hoover over the coloured box. The different types of colored boxes are explained below.
Methods:
If the submitter of the data provided some specific remarks, you can read these in the last column by hoovering over the word 'remarks'.
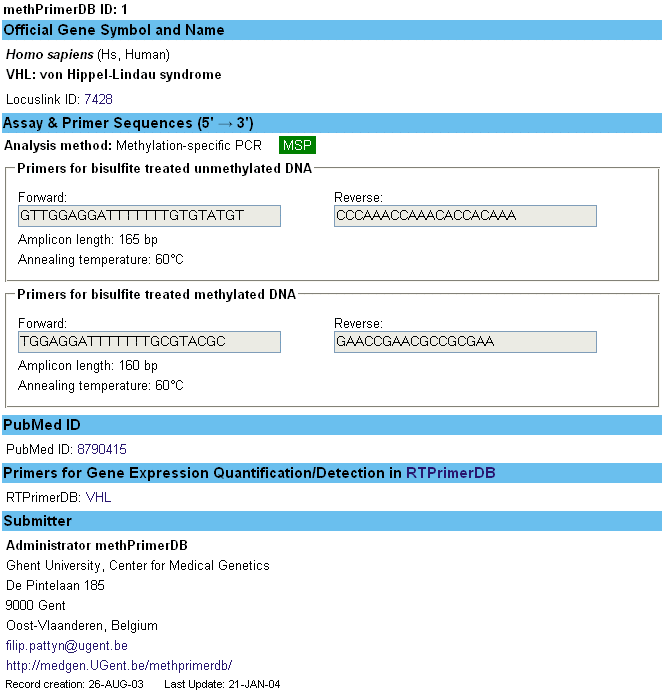
Assay report
Every assay report contains all the available data about a certain primer set.
The data is split in the following blocks:
- The methPrimerDB ID is displayed in the first column.
- Official/Interim Gene Symbol and Name: Depending on the revision state of the gene, the gene name or gene symbol is marked as 'official' or 'interim'. Reviewed genes get an official name from a nomenclature committee.
Here, the latin name, the abbreviation and the english name of the organism are shown together with the gene symbol, gene name, and possible alias symbols. The next line contains the Entrez Gene ID, which is linked to NCBI's Entrez Gene database.
- Assay & Primer Sequences (5' → 3'): The DNA methylation analysis method.
The primer sequences, the annealing temperature and the amplicon length.
- PubMed ID: An optional field with a link to an article in which the submitted primers/probes are described.
- Submitter's Remarks: An optional field with additional information provided by the submitter.
- Submitter: Personal information about the submitter: name, address, e-mail address (optional), web site (optional).
- The last line contains the creation date and the date of the last (submitter's) update of the primer/probe record.
Underneath, the assay report of the first record in the database, for the human gene VHL, is shown.
| How to register? |
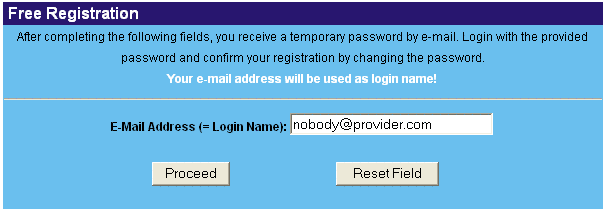
If you want to submit your own primer sequences to the DNA methylation Analysis PCR Primer Database, you have to register to obtain data insertion rights. Click on 'Registration' > 'Register' and complete the following form. Use your professional email adress as login.
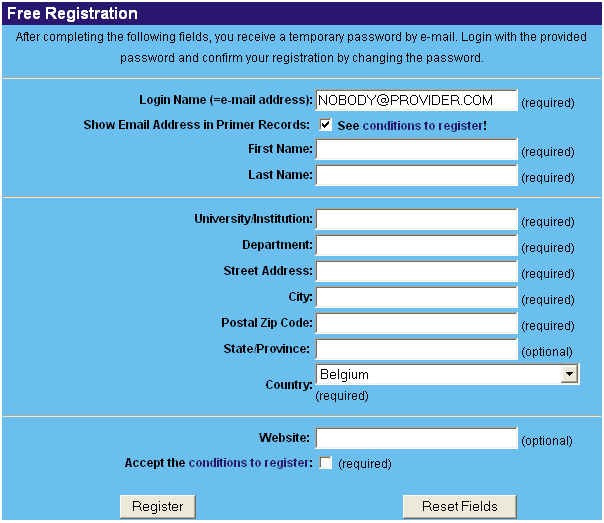
Read the 'conditions to register' carefully and decide if you want to make your email adress publicly available or not. Fill in all the required fields to make your registration complete.
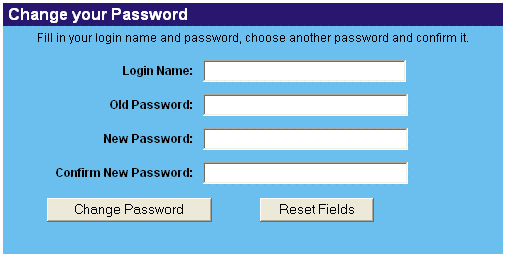
After registration, you will receive a temporary password via e-mail. You cannot submit data before you changed your password. To do this, follow the link provided in the e-mail or click on 'Registration' > 'Change Password' and fill in your login name, your temporary password, your new password and confirm by typing the new password a second time.
| How to submit data? |
| How to submit large amounts of data? |
| Which fields need special attention for data submission? |
This public database project aims at reliable methylation analysis PCR primer information. Therefore, it's very important that submitters accurately complete all the necessary data fields.
Some fields are required and are marked with '(required)', others are optional. The following list indicates the fields for which special attention is needed:
- Official Gene Name: The official abbreviation (not aliases) used by a Nomenclature Committee. See the Links Page to check which organism specific database you should consult to find the official gene name.
- Forward Primer & Reverse Primer: Type/paste the primer sequences (from 5' to 3'), without any other information.
Only official IUB/IUPAC abbreviations are allowed:
- A --> adenosine
- C --> cytidine
- G --> guanine
- T --> thymidine
- U --> uridine
- R --> G A (purine)
- Y --> T C (pyrimidine)
- K --> G T (keto)
- M --> A C (amino)
- S --> G C (strong)
- W --> A T (weak)
- B --> G T C
- D --> G A T
- H --> A C T
- V --> G C A
- N --> A G C T (any)
- PubMed-ID: The PubMed identifier of an article in which the primers/probes are described for the first time. This type of information makes the primer/probe data more trustworthy.
- Remarks: Type here some additional/helpful information about the primer set for other users.
| How to change your registration properties? |
| How to construct a link to a specific record? |
Use the following syntax:
http://methprimerdb.cmgg.be/methprimerdb/assay_report.php?methPrimerDB_ID=<methPrimerDB ID>
Example: http://medgen.ugent.be/methprimerdb/assay_report.php?methPrimerDB_ID=1
| How to quote/cite the use of primers from the methPrimerDB database? |
| What's the software and hardware behind this database? |
The methPrimerDB database is managed by an Oracle 9i relational database management system.
The web application is based on PHP scripts using the Oracle Call Interface (OCI).
The complete application runs on a nginx/1.24.0 server at methprimerdb.cmgg.be (157.193.244.57)
|
Ghent University, Faculty of Medicine and Health Sciences, Centre for Medical Genetics 1K5, De Pintelaan 185, B-9000 Gent, Belgium Tel.: +32 9 240 55 18 - fax: +32 9 240 49 70 - Webmaster - Disclaimer & copyright |

|